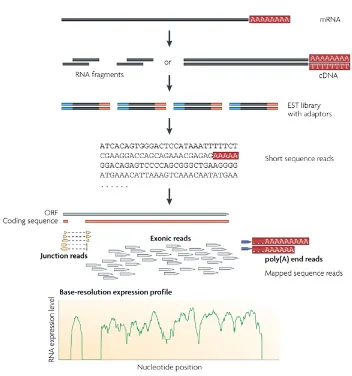
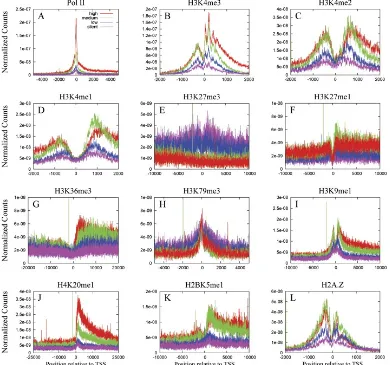
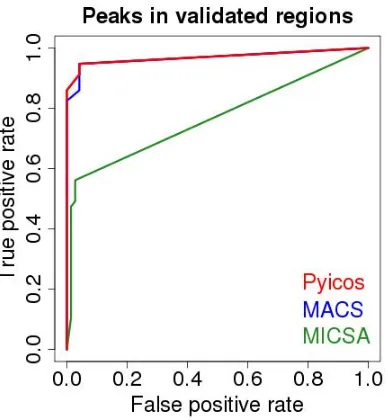
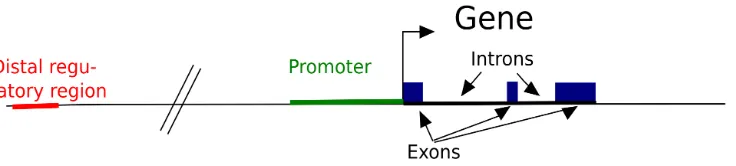
Elucidating mechanisms of gene regulation. Integration of high-throughput sequencing data for studying the epigenome
Texto completo
Figure
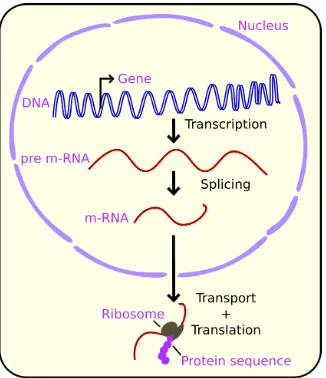
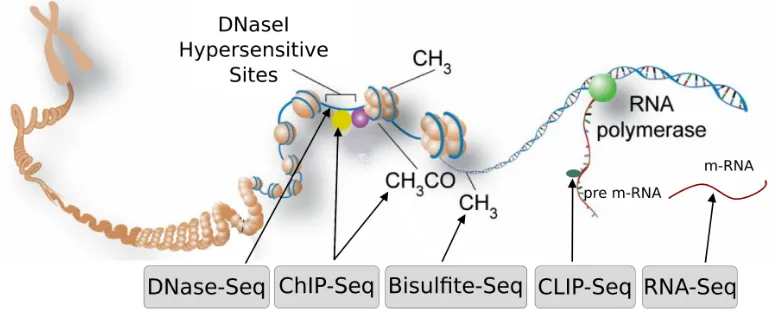
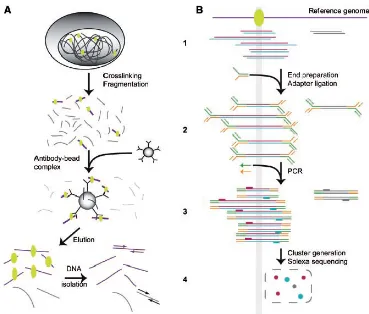
Documento similar
Therefore, utilizing 319 high-throughput sequencing of 16S rRNA (region V3-V4) may allow for a complete understanding 320 of the variations in the structure and abundance
The obtained nucleotide sequence was used to develop RT-PCR, RT-PCR with SYBR and a TaqMan probe, and RT-LAMP for the fast and specific detection and quantification of xufa yellow
On the other hand, extrapolation methods constitute one of the most efficient classes of schemes for the numerical integration of the second order differential equation (1.1) when
The impossibility of solving the atomic structure of full-length TH has been for years a limitation to fully understand the regulation mechanisms of this enzyme. This
11 Deciphering the role of protein-protein interaction networks in the functional profiling of high-throughput experiments 81 11.1 Ppi networks in Gene Ontology
Lara gracias por tu amistad y tus buenos consejos, León es imposible aburrirse contigo, gracias por ser un buen amigo; Ana y Sandra, muchas gracias por vuestra compañía
In order to check for any “activity amount”- related effects, we further analyzed the samples by splitting runner and high runner (Fig. The results seem to indicate a
The relationship between the coverage obtained using IS-seq (reads per RFLP band) and the difference be- tween observed and expected sites showed that our method was prone to