Availableonlineatwww.sciencedirect.com
Revista
Mexicana
de
Biodiversidad
www.ib.unam.mx/revista/ RevistaMexicanadeBiodiversidad87(2016)18–28
Taxonomy
and
systematics
Assessment
of
non-cultured
aquatic
fungal
diversity
from
different
habitats
in
Mexico
Estimación
de
la
diversidad
de
hongos
acuáticos
no-cultivables
de
diferentes
hábitats
en
México
Brenda
Valderrama
a,
Guadalupe
Paredes-Valdez
a,
Rocío
Rodríguez
b,
Cynthia
Romero-Guido
b,
Fernando
Martínez
b,
Julio
Martínez-Romero
c,
Saúl
Guerrero-Galván
d,
Alberto
Mendoza-Herrera
e,
Jorge
Luis
Folch-Mallol
b,∗aInstitutodeBiotecnología,UniversidadNacionalAutónomadeMéxico,AvenidaUniversidad2001,Col.Chamilpa,62210Cuernavaca,Morelos,Mexico bCentrodeInvestigaciónenBiotecnología,UniversidadAutónomadelEstadodeMorelos,AvenidaUniversidad1001,Col.Chamilpa,62209Cuernavaca,
Morelos,Mexico
cCentrodeCienciasGenómicas,UniversidadNacionalAutónomadeMéxico,AvenidaUniversidads/n,Col.Chamilpa,62210Cuernavaca,Morelos,Mexico dCentroUniversitariodelaCosta,UniversidaddeGuadalajara,AvenidaUniversidad203,DelegaciónIxtapa,48280PuertoVallarta,Jalisco,Mexico eCentrodeBiotecnologíaGenómica,InstitutoPolitécnicoNacional,BoulevarddelMaestros/nesq.ElíasPi˜na,Col.NarcisoMendoza,88710Reynosa,
Tamaulipas,Mexico
Received20August2013;accepted27September2015 Availableonline28February2016
Abstract
WiththeaimtoexplorethediversityofaquaticfungiinMexicowepresentaninvestigationusingafragmentofthe18SribosomalDNAas amolecularmarkerobtainedfromdifferentwaterbodies(marine,brackishandfreshwater).RibosomalgenefragmentswereobtainedbyDNA amplification,theresultingsequenceswerecomparedusingmultiplealignmentsagainstacollectionofclassifiedreferencefungalsequencesand thensubjectedtophylogeneticclusteringallowingtheidentificationandclassificationofDNAsequencesfromenvironmentalisolatesasfungal downtothefamilylevel,providedenoughreferencesequencewereavailable.Fromourensembleof2,020sequencesidentifiedasfungal,23.8% wereclassifiedatthefamilylevel,48.5%attheorderlevel,13%attheclass/subphylumleveland14.7%ofthesequences(allfromthesame site)couldnotbeunambiguouslypositionedinanyofourreferencefungalgroupsbutwerecloselyrelatedtouncultivatedmarinefungi.The mostfrequentlyrecoveredphylumwasAscomycota(89.1%),followedbyChytridiomycota(8.1%),Basidiomycota(2.8%)andMucoromycotina (1.3%).
AllRightsReserved©2015UniversidadNacionalAutónomadeMéxico,InstitutodeBiología.Thisisanopenaccessitemdistributedunderthe CreativeCommonsCCLicenseBY-NC-ND4.0.
Keywords: Aquatichabitats;Fungi;Taxonomicclassification;Fungalpopulations;18SribosomalDNA
Resumen
Con lafinalidad deexplorarladiversidad dehongosacuáticosen México,se presentauna investigaciónusando unfragmento del ADN ribosomal18Scomounmarcadormolecularobtenidodemuestrasdecuerposacuáticoscondiferentescaracterísticas(marino,salobreydulce). LosfragmentosdelosgenesribosomalesseobtuvieronmediantelaamplificacióndeADN,lassecuenciasresultantessecompararonmediante alineamientosmúltiplesconunaseleccióndesecuenciasdehongoscomoreferenciayposteriormenteseanalizaronfilogenéticamente,permitiendo laidentificaciónyclasificacióndesecuenciasdeADNprovenientesdeaisladosambientaleshastalacategoríadefamilia,cuandohubosuficientes secuenciasdisponibles.Delas2,020secuenciasidentificadascomohongos,un23.8%seclasificaroncomofamilia,un48.5%comoorden,un13%
∗Correspondingauthor.
E-mailaddress:jordi@uaem.mx(J.L.Folch-Mallol).
PeerReviewundertheresponsibilityofUniversidadNacionalAutónomadeMéxico.
http://dx.doi.org/10.1016/j.rmb.2016.01.013
comoclase/subphylumyun14.7%delassecuencias(todasdelmismolugar)nopudieronsercolocadasinequívocamenteenalgunodelosgrupos dehongosquesetomaroncomoreferencia,peroseencontraronmuycercanamenterelacionadasahongosmarinosnocultivables.Elphylummás representadofueAscomycota(89.1%),seguidodeChytridiomycota(8.1%),Basidiomycota(2.8%)yMucoromycotina(1.3%).
DerechosReservados©2015UniversidadNacionalAutónomadeMéxico,InstitutodeBiología.Esteesunartículodeaccesoabiertodistribuido bajolostérminosdelaLicenciaCreativeCommonsCCBY-NC-ND4.0.
Palabrasclave: Hábitatsacuáticos;Hongos;Clasificacióntaxonómica;Poblacionesfúngicas;ADNribosomal18S
Introduction
Currentestimatesoffungaldiversitybasedontheplant:fungi ratio found in countries where both populations are suffi-cientlywellstudiedsuggesttheexistenceof1.5millionspecies (Hawksworth,1991,2001;Mueller&Schmit,2007).Thestudy offungaldiversityisimportantbecausefungiaredecomposers oforganicmatterandcompriseamajorproportionofmicrobial biomass.Recently,globalclimatechangeandthebetterknown roleoffungiinbiogeochemicalcycleshaveenforcedthe impor-tanceofstudyingfungaldiversity(Chapinetal.,2000;Wardle &Giller,1996).
Studiesoffungaldiversityhavebeenlimitedbythelackof appropriatemicrobiologicalmethods(Kimura,2006;Torsvik& Ovreas,2002).The applicationof molecularapproachessuch asextracting,cloningandamplifyingDNAfromenvironmental samplescurrentlyallowsustoexplorebiodiversitywithoutthe needofculturing.Inthisregard,18SribosomalDNA(rDNA) sequenceshavebeenextensivelyusedtoexplorefungal diver-sity (Hunt, Boddy, Randerson, & Rogers, 2004; Le Calvez, Burgaud,Mahé,Barbier,&Vandenkoornhuyse,2009;Monchy etal.,2011;Piquet,Bolhuis,Meredith,&Buma,2011)andmany specificprimershavebeendesignedforthispurpose(Borneman &Hartin,2000;Moon-vander Staay,De Wachter,&Vaulot, 2001;Vainio&Hantula,2000).Thecapacityoftheseprimers to reveal fungal diversity in environmental samples is based ontheirspecificityinpreferentiallypriming fungalsequences andalsotheirabilitytorepresentallfungalphylaatthe same time(Anderson,Campbell,&Prosser,2003;Huntetal.,2004). Moleculartools,including18SrDNAsequenceanalysis,have beenusedrecentlytore-definefungaltaxonomybasedon multi-locusphylogeneticanalyses(Hibbettetal.,2007;Jamesetal., 2006).Asaconsequence,ourviewoftraditionalfungalgroups haschangeddrastically.
Ifterrestrialfungiare stilllargelyunder-described,aquatic fungiareevenlesswellknown.Mostofthecultivatedaquatic speciesbelongtotheChytridiomycotaandAscomycotaphyla (Mueller & Schmit, 2007; Mueller, Bills, & Foster, 2004; Sheareretal.,2007)andmanyfungal-relatedmicrobes belong-ingtothestraminipiles (oomycetesandhyphochytriomycetes inparticular) havebeen described (Mueller etal., 2004; Van der Auwera et al., 1995). In Mexico, an important effort to explorefungaldiversityhasbeenmade(Guzmán,1998). Explo-rationof aquaticfungaldiversity hasalsobeen conductedby traditionalmethodsisolatingfungifromfreshwaterandmarine environments(González&Chavarría,2005;González,Hanlin, Herrera, & Ulloa, 2000; González, Hanlin, & Ulloa, 2001; Heredia,Reyes,Arias,Mena-Portales,&MercadoSierra,2004).
In marine environments, ascomycetes and mitosporic fungi weremainlyfound,althoughonebasidiomycetewasreported (Gonzálezetal.,2001).
To explorethe diversityofaquaticfungiinMexicoandto demonstratethepotentialofaclassificationsystembasedona singlemolecularmarker,wepresentaninvestigationofdifferent waterbodies(marine,brackishandfreshwater)usingafragment of 18S rDNA sequences andthe results of our phylogenetic clusteringapproach.
Materialsandmethods
Descriptionofthesampledlocations
Zempoala, Morelos (fresh water, 19◦0120N, 99◦1620W). The Zempoala Lagoons comprises 7 pris-tine water bodies located in a protectedpark in the state of Morelos at 2670–3686masl. The area is surrounded by a temperateforestofpines,firsandoaks.Sampleswereobtained fromoneofthepermanentlagoons.
Carboneras, Tamaulipas (brackish water, 24◦3741.88N, 97◦4259.19W). Fishery and leisure town located at 58km from San Fernando inthe state of Tamaulipas. It belongs to thecentralsectionofLagunaMadre.
Mezquital, Tamaulipas (sea water, 25◦1455.70N, 97◦3105.54W).Locatedintheeasternsideofthewiderpart ofLagunaMadre,itisconnectedtotheGulfofMexicothrough anartificialnavigationchannel.
Media Luna, Tamaulipas(brackishwater, 25◦0947.64N, 97◦4016.35W).Locatedatthewesternsideofthewiderpart of LagunaMadre,andduetopoor roadconditionsandtothe absenceoflargesettlements,thisisoneofthelessspoiledareas. ThedistancebetweentheMezquitalandMediaLunasampling placesisapproximately12km.
El Rabón, Tamaulipas (hypersaline sea water,
25◦2623.68N, 97◦2434.79W). At the northern end of LagunaMadre, thisareahassufferedserious transformations due to human activity and had become dry. Recently, the wetlandshavebeenrestoredbypumpinginseawater.
Carpintero. Tamaulipas (fresh water, 22◦1401.12N, 97◦5120.67W). Belonging to the Pánuco River basin and located in a protectednatural parkcovering 7ha, Carpintero Lagooniscurrentlyusedforfishandcrocodilebreedinggrounds. InspiteoftheurbanlocationofthesiteintheCityofTampico itisrelativelyunspoiled.
20 B.Valderramaetal./RevistaMexicanadeBiodiversidad87(2016)18–28
abovesealevel.Withanaveragevolumeof500millionm3itis usedforaquaticsportsandsportfishing.
Bahía de Banderas, Jalisco (sea water, 20◦3858.93N, 105◦2451.94W).Locatedintheborderbetweenthestatesof NayaritandJaliscointhePacificCoast,itisthelargestbayin Mexico with asurface of 773km2. The Ameca Rivermouth dividesthebay,whichhasahighpopulationdensitybasedon PuertoVallarta.
Cruz de Huanacaxtle, Nayarit(sea water, 20◦4412.96N, 105◦2317.53W).Acoastalsitewithlowanthropogenicimpact locatedinthe northendofBahía deBanderasinthestate of Nayarit.ThedistancebetweentheBahíadeBanderasandCruz deHuanacaxtlesampledsitesisapproximately15km.
Zacapulco,Chiapas(mangroveswampwater,15◦0407N, 92◦4520W).Unspoiledmangrovearealocated200km north-westof theborderofthe stateofChiapaswithGuatemalaon the PacificOcean coastline.At14masl,theestuarine salinity oscillateswiththetides.Theperennialvegetationprovides cov-erageforthehabitatofmanyaquaticbirds.Thereisnoseasonal daytimechangeandtemperaturesvarybetween25and35◦C.
Santa Catarina, Querétaro (fresh water, 20◦4730.6N, 100◦2701.66W).An artificialwaterreservoirlocated25km northwestofthecityofQuerétarointhestateofQuerétaroat 2035masl.
Samplecollection
We decided to collect samples from water instead from organicmattertorecoverawiderselectionofthefungal popu-lationineachsite,notonlyofthosedirectlyinvolvedindecay. Samples were collected from 0.5m below the water surface using acleanandsurfacesterilizedcontainer.Typicalsample volumeswere20L.Thewholesamplewaspassedthrough3 lay-ersofsterilecheeseclothandafterwardsfilteredthrough5m PVDFmembranes(Millipore).Thebiomasslayerwasscrapped withaspatula fromthe membranes,washedin3–5ml ofthe samewaterandcentrifugedin3differentEppendorftubesfor replica analysis. Final biomass volumes were of 0.1–0.3ml. Pelletswerefrozenat−20◦Cuntilextraction.
DNAextraction
The whole pellet was processed andtotal DNA extracted usingtheUltraCleanSoilDNAKit(MoBio,Carlsbad,USA)in accordancetothemanufacturer’sinstructions.TotalDNAwas analyzedforintegritybyagarosegelelectrophoresis.
RibosomalDNAamplification
TotalDNAaliquotswereamplified usingprimers nu-SSU-0817andnu-SSU-1536(Borneman&Hartin,2000),yieldinga primary product of approximately 750bp.Reaction mixtures contained 2.5mM MgCl2, 1mM dNTPs mix, 5pM of each primer,50ngoftotalDNAastemplate,1Xreactionbufferand 5UofTaqpolymerase(Altaenzymes,Alberta,Canada). Reac-tionmixturesweresubjectedtoaninitialdenaturationstepof 5minat95◦Cfollowedby30amplificationcycles(30sat95◦C,
30sat55◦C,45sat72◦C)andafinalextensionstepof2min at72◦C.AmplificationswereperformedinaPCRsprint ther-malcycler(ThermoElectronCorporation,USA).Theresultant fragments were separated by agarosegel electrophoresis, the amplifiedfragmentwasvisualizedbyethidiumbromide stain-ing,excisedandpurifiedwiththeQIAquickgelextractionkit (QIAGEN,Hilden,Germany).
Libraryconstruction
AmplifiedfragmentsweredirectlyclonedinthepGemT-easy vector(Promega,Madison,USA)andtransformedby electro-poration into Escherichiacoli strain DH5␣.Insert-containing clonesweredetectedbythelackofcolorationinthepresence of X-galandIPTGandpickedinfreshplates.Plasmidsfrom 6 randomly selectedclones from each library were extracted byalkalinelysisandsequencedforinsertverification.Inthose caseswheretheamplificationproductcouldnotbedetectedin atleast4outofthe6clonesorwhenthesequencedinsertswere not offungalribosomalgenes,the procedurewas repeatedto obtainabetteryieldorquality.
DNAsequencing
Agroupof384randomlyselectedclonesfromeachlibrary wassequencedinsetsof496-wellplates.Colonieswerepicked andplasmidDNAextractedbyalkalinelysis.Theconcentration andintegrityofisolatedDNAwereverifiedbyagarosegel elec-trophoresisandsequencedwiththeT7primerintheSequencing UnitoftheCentrodeCienciasGenómicas(UNAM).
Controlexperiments
GenomicDNA ofdifferent organismswasused as control beforesampleamplification.Fromprokaryoticsources:E.coli,
RhizobiummelilotiandSpirulinamaxima.Fromfungalsources:
Aspergillusnidulans,Debaryomyceshansenii,Yarrowia lipoly-tica,Saccharomyces cerevisiae, Schizophyllumcommune and
Bjerkandera adusta. For an arthropod source we used DNA fromCentruroideslimpidusandfromplantsources,Arabidopsis thalianaandPhaseolusvulgaris.Inallcasesthesampleswere kindlyprovidedbycolleaguesattheInstitutodeBiotecnología (UNAM) and the Centro de Investigación en Biotecnología (UAEM).
Sequenceanalysis
Table1
Taxonomicclassificationofreferencesequencesusedinthiswork.
Phylum Class/Subphylum Order Family Species GI
Ascomycota Saccharomycetes Saccharomycetales Mitosporic Saccharomycetales
CandidasakestrainJCM8894 4586748
Candidafluviatilis 4586709
CandidamembranifaciensstrainW14-3 124494629
Saccharomycetaceae Kazachstaniasinensis 114050511
Kluyveromyceshubeiensis 33114591
Saccharomycescerevisiae 270308944
Eurotiomycetes Onygenales Onygenaceae Castanedomycesaustraliensis 21732245
Mycocaliciales Mycocaliciaceae Chaenothecopsissavonica 2804615
Chaetothyriales Incertaesedis Coniosporiumsp.MA4597 66990818
Eurotiales Trichocomaceae Penicilliumsp.EnrichmentculturecloneNJ-F4 270311611
Aspergillussp.Z3b 151384867
AspergillusunguisstrainF3000054 120431388
Leotiomycetes Helotiales Bulgariaceae BulgariainquinansisloteAFTOL-ID916 91841147
Dermateaceae PeziculacarpineaisolateAFTOL-ID938 91841226
Dothideomycetes Capnodiales Davidiellaceae MycosphaerellatassianastrainTS01 238734423
Capnodiaceae Leptoxyphiumsp.MUCL43740 50726934
Pleosporales Pleosporaceae Alternariasp.enrichmentculturecloneNJ-F7 270311614
Glyphiumelatum 17104830
Phomasp.CCF3818 213876689
Incertaesedis Norrliniapeltigericola 56555555
Sordariomycetes Xylariales MitosporicXylariales Dicymaolivacea 13661088
Hypocreales Mitosporic
Hypocreales
Fusariumoxysporum 291482357
Ophiocordycipitaceae Hirsutellacitriformis 11125693
Incertaesedis PutativePaecilomycessp.080834 89112992
Diaporthales Valsaceae ValsellasalicisisolateAFTOL-ID2132 112785209
Cryphonectriaceae ChrysoporthecubensisisolateAFTOL-ID2122 112785199
Basidiomycota Ustilaginomycetes Ustilaginales Ustilaginaceae Pseudozymasp.JCC20718S 77167276
Agaromycetes Polyporales Polyporaceae Coriolopsisbyrsina 288557592
Agaricales Cyphellaceae Radulomyceshiemalisisolate5444a 116687716
Auriculariales Auriculariaceae AuriculariaceaecloneAmb18S699 134022019
Russulales Peniophoraceae Peniophoranuda 2576440
Tremellomycetes Tremellales Tremellaceae Cryptococcusvishniacii 7262452
Filobasidiales Filobasidiaceae Filobasidiumglobisporum 21326776
Cystofilobasidiales Cystofilobasidiaceae Cystofilobasidiuminfirmominiatumisolate AFTOL-ID1888
109289344
Microbotrymycetes Leucosporidiales Nonidentified LeucosporidiumscottiisolateAFTOL-ID718 51859977 Sporidiobolales Mitosporic
Sporidiobolales
RhodotorulaglutinisAFTOL-ID720 111283841
Blastocladiomycota Blastocladiomycetes Blastocladiales Catenariaceae Catenomycessp.JEL342isolateAFTOL-ID47 49066429 Chytridiomycota Chytridiomycetes Chytridiales Chytridiaceae BlyttiomyceshelicusisolateAFTOL-ID2006 108744678
Chytriomycessp.JEL378isolateAFTOL-ID 1532
108744670
Chytriomycetaceae EntophlyctishelioformisisolateAFTOL-ID40 49066425
Entophlyctissp.JEL174isolateAFTOL-ID38 49066423 Cladochytriaceae PolychitriumaggregatumstrainJEL109 47132215 Rhizophlyctidiales Rhizophlyctidaceae RhizoplhyctisroseaisolateAFTOL-ID43 490664428
RhizophydiumelyenseisolateAFTOL-ID693 108744666
TriparticalcararcticumisolateAFTOL-ID696 108744667 Fungiincertae
sedis
Mucoromycotina Mucorales Mucoraceae MucorplumbeusstrainUPSC1492 33334392
Thamnidiaceae Backusellactenidia 11078007
Fungi/metazoan incertaesedis
Protozoa Eccrinales Eccrinaceae EccrinidusflexilisisolateSPA11C45 50083273
Rozellida Rozelliidae RozellaallomycisisolateAFTOL-ID297 49066437
Rozellasp.JEL347isolateAFTOL-ID16 47132211
Ichthyosporeae Nonidentified Nonidentified Ichthyoponidasp.LKM51 3894141
Ichtyophonida Nonidentified Anurofecarichardsi 4322029
Nonidentified Choanoflagellida Acanthoecidae Stephanoecadiplocostata 37359232
Salpingoecidae Lagenoecasp.antarctica 120407515
Alveolata Dinophyceae Nonidentified Unclassified
Dynophyceae
22 B.Valderramaetal./RevistaMexicanadeBiodiversidad87(2016)18–28
fungiso wedecided toleave allambiguoussequencesinthe set for further analysis. Approved sets were automatically aligned using the Clustal W algorithm (Larkin et al., 2007) containedinMegaversion4(Tamura,Dudley,Nei,&Kumar, 2007).Mostofthesequencealignedinthefirstround,although optimizationof the surroundingsof variable regionsrequired manualalignment.Vectorbornefragmentswereremovedafter alignment.Asetofribosomalsequencesfromwell-identified organisms(Table1)wasaddedtoeach alignmentforinternal referenceandthegroupwasre-alignedandmanuallyoptimized withClustalW.Wesometimesdetectedgroupsofexperimental sequences that did not cluster with reference sequences but withinthemselves.Inthesecasesweidentifiedeachoneofthe sequences by looking for the closestmatch in the databases usingtheBLASTtool.Forsomesequences,theclosestmatch resulted to be an entry from a characterized species but in othercaseswe recoveredentriesfromenvironmentalsurveys. Phylogenies were reinforced by including sequences from characterizedspeciestothereferencesequencelistand, some-times,sequencesfromunculturedsources(Table2).Sequence clusteringwasperformedusingtheNeighborJoiningalgorithm (Saitou&Nei,1987)containedinMegaversion4.
Results
Amplificationoffungal18SribosomalDNAfromreference isolates
Control genomic DNA from different sources was tested foramplificationwithprimersnu-SSU-0817andnu-SSU-1536. Templates from non-fungal sources were unable to support amplification while fungal sources specifically amplified the internalfragmentof18SrDNA(datanotshown).In5outof6 fungalsamples(A.nidulans,D.hansenii,Y.lipolytica,S. cere-visiae,andB.adusta)thesizeoftheamplifiedproductmatched theexpected762bp(datanotshown).Sequencedataqualitywas assessedbysequencingacontrollibrarygeneratedby amplifi-cationof18SrDNAfromS.cerevisiae.
Identificationandphylogeneticanalysisofnu-SSU-0817 andnu-SSU-1536amplificationlibraries
TotalDNAfromwatersamplesfrom11differentsites(see Section ‘Materialsandmethods’)wasisolated, afragment of the 18S rDNA amplified and cloned in genetic libraries for individual clonesequencing.In thoserarecaseswhererDNA
Table2
Non-culturedreferencesequencesusedinthiswork.
Definition CloneID GInumber ClosestmatchbyBLAST
Unculturedfungi CloneBAQA254 20377933 Phaeopleosporaeugeniicola(Ascomycota)
UnculturedbasidiomycetecloneH18E128 149786618 Filobasidiumglobisporum(Basidiomycota) UnculturedbasidiomycetecloneMV2E89 149786738 Cryptococcusvishniacci(Basidiomycota) UnculturedbasidiomycetecloneMV5EEF18 149786752 Pseudozymasp.(Basidiomycota) UnculturedbasidiomycetecloneLC235EP14 95115857 Pseudozymasp.(Basidiomycota)
Unculturedascomycete 27530772 Davidiellatassiana(Ascomycota)
UnculturedascomycetecloneLC234EP18 95115853 Bulgariainquinans(Ascomycota)
Unculturedascomyceteisolate 21902393 Phomasp.(Ascomycota)
Unculturedrhizosphereclone 23504803 Mucorplumbeus(Mucoromycotina)
UnculturedChytridiomycotacloneMV5E291 149786790 Rhizophydiumelyensis(Chytridiomycota)
CloneCCW24 29423782 Triparticalcararcticum(Chytridiomycota)
CloneCCW48 27802600 Entophlyctisconfervae-glomeratae(Chytridiomycota)
Clonecontrol46 151413777 Rozellasp.JEL347(Chytridiomycota)
CloneRBfung138 90904231 Candidasp.Y6EG-2010(Ascomycota)
CloneWIM48 113926798 Basidiobolushaptosporus(Fungiincertaesedis)
CloneF47(S2) 86604435 Preussialignicola(Ascomycota)
CloneNAMAKO-37 114217391 Basidiobolushaptosporus(Fungiincertaesedis)
CloneSSRPD64 126033366 Phaeophleosporaeugeniicola(Ascomycota)
CloneZeuk2 59709949 Phaeophleosporaeugeniicola(Ascomycota)
Unculturedeukaryotes Clone051025T2S4WTSDP12094 223030789 Cryothecomonaslongipes(Cryomonadida)
Clone18BR20 124541005 Paracalanusaculeatus(Copepoda)
CloneSCM15C21 56182170 Pentapharsodinumtyrrhenicum(Dinoflagellate)
CloneSCM27C27 50541716 Acartialongiremis(Copepoda)
CloneSCM28C135 56182295 Diaphanoecagrandis(Choanoflagellidae)
CloneSCM37C13 50541727 Pantachogonhaeckeli(Cnidaria)
CloneSCM38C38 50541719 Paracalanusparvus(Copepoda)
CloneSCM38C41 56182194 Ichthyodiniumchabelardi(Alveolata)
CloneSCM38C62 56182178 Dinophyceaesp.CCMP1878(Dinoflagellate)
CloneSCM38C9 50541718 Paracalanusparvus(Copepoda)
CloneSSRPB26 126033229 Gymnodiniumaureolum(Alveolata)
CloneTAGIRI-8 67624905 Gymnodiniumbeii(Alveolata)
CloneCYSGM-24 133778655 Amastigomonasmutabilis(Apusozoa)
IsolateE3 30144455 Duboscquellasp.Hamana/2003(Alveolata)
CloneMB04.31 146157556 Oithonasimilis(Copepoda)
sequences from phylogenetic groups other than fungi were recovered,thesewereremovedfromthecollectionbefore clus-tering. The only exception to this behavior was the sample from Carpintero lagoon, where the 146 sequences recovered weremoresimilartoreferencesequencesfromdinoflagellates thanfromfungi.Thissetofsequenceswasremovedfromour analysis.
Clusteringanalysisofthelibraries
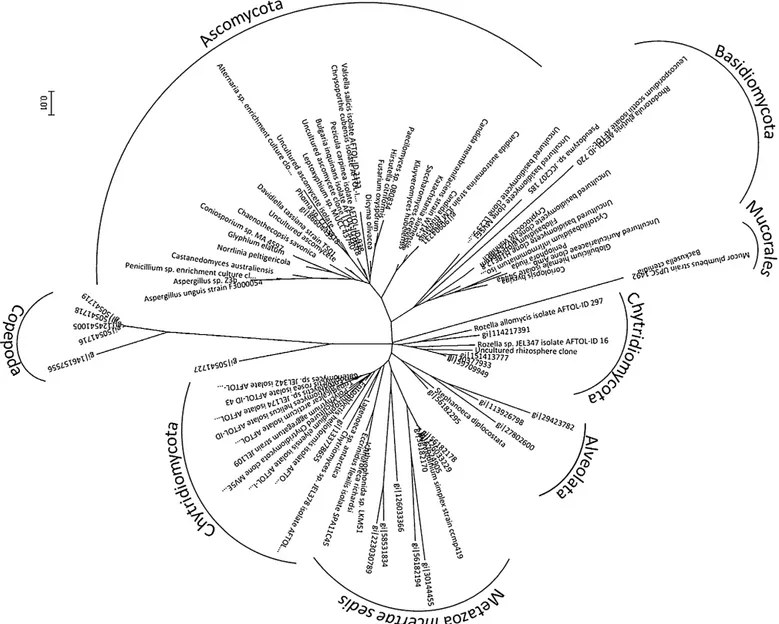
Wetestedtheabilityofourmolecularmarker,afragmentof the 18rDNA gene containing regionsV4–V8, toreconstruct a clustering profile consistent withthe taxonomic classifica-tionusingourensembleofreferencesequences(Fig.1).Once therobustnessoftheclusteringmethodwasdemonstrated,the environmental sequences from 10 of our 11 libraries (after removingthesamplesfromCarpinteroLagoonforthereasons described above) were organizedby site along withthe fun-galreferencesequencesasdescribedinSection‘Materialsand
methods’. Serialreconstructions of the 10libraries were per-formedusingthemanuallyoptimizedmultiplealignmentfrom each site, whichincluded the recovered referencesequences, untilarobustandinformativephylogramwasobtained.Based onthesephylograms,weidentifiedthemostprobabletaxonomic classificationoftheenvironmentalsequencesbytheirclustering withreferencesequences.
Bythemethodologydescribedabove,wewereableto iden-tify529oftheenvironmentalsequencesatthefamilylevel,1077 attheorderleveland288attheClass/Subphylumlevel.Only 326sequences,allfromthesamesite,didnotgroupwithany ref-erencesequencebuttouncultivatedclonesfrommarinesources (Table3).ThemostabundantphylumfoundwasAscomycota (1458sequences)(Fig.2),followedbyChytridiomycota(133 sequences)(Fig.3).Sequencesbelongingtophylum Basidiomy-cota(45sequences)(Fig.4)andtosubphylumMucoromycotina (21 sequences)wereseldom recovered as wellas non-fungal sequencesbelongingtoChoanoflagellidaeandEccrinales (29 sequences)(Fig.5).
24
B.
V
alderr
ama
et
al.
/
Re
vista
Me
xicana
de
Biodiver
sidad
87
(2016)
18–28
Table3
Identificationofenvironmentalsequences.
Sampledsites
Phylum Class/Subphylum Order Family Genus Zempoala Carboneras Mezquital Cruz Huanacaxtle
Vicente Guerrero
Media Luna
El rabón
Zacapulco Santa Catarina
Bahía Banderas
Totalby genus
Saccharomycetes (363)
Saccharomycetales Mitosporic Saccharomycetal
Candida 63 6 69
Nonidentified Nonidentified 9 213 71 293
Nonidentified Nonidentified Nonidentified 1 1
Nonidentified(260) Nonidentified Nonidentified Nonidentified 7 253 260
Leotiomycetes(3) Heliotiales Dermateaceae Pezicula 3 3
Ascomycota (1279)
Eurotiomycetes (18)
Eurotiales Trichocomaceae Aspergillus 1 1 5 7
Chaetothyriales Nonidentified Nonidentified 6 6
Verrucarriales Verrucariaceae Norrlinia 1 1
Nonidentified Nonidentified Nonidentified 4 4
Sordariomycetes (630)
Hypocreales Nonidentified Nonidentified 1 10 109 120
Diaporthales Cryphonectriaceae Chysoporthe 8 1 9
Hypocreales Mitosporic Clavicipitaceae
Paecilomyces 24 12 61 11 234 26 368
Nonidentified Nonidentified 133 133
Euascomycetes(1) Pleosporales Pleosporaceae Phoma 1 1
Dotyideomycetes(1) Capnodiales Davidiellaceae Davidiella 1 1
Nonidentified(3) Nonidentified Nonidentified Nonidentified 2 1 3
Mucorales(27) Nonidentified(27) Nonidentified Nonidentified Nonidentified 4 4 19 27
Chytridiomycota (299)
Nonidentified Nonidentified Nonidentified 3 3
Nonidentified Nonidentified 35 35
Spizellomycetales Spizellomyceteaceae Tripalcalcar 6 6
Spizellomycetales incertaesedis
Rozella 13 179 4 23 1 5 225
Chytridiomycetes (299)
Chytridiales Endochytriaceae Nonidentified 1 1
Nonidentified Nonidentified 4 4
Chytridiaceae Blyttiomyces 10 10
Chytriomyces 1 3 4
Cladochytriaceae Polychytrium 4 4
Rhizophydiales Rhizophydiaceae Rhizophydium 4 2 1 7
Basidiomycota (40)
Tremellomycetes (9)
Filobasidiales Nonidentified Nonidentified 3 3
Filobasidieaceae Nonidentified 2 1 3
Cystofilobasidales Cystofilobasidiaceae Nonidentified 3 3
Agaromycetes(14) Polyporales Nonidentified Nonidentified 1 13 14
Pucciniomycotina(2) Microbotrimycetes Leucosporidiales Leucosporidium 2 2
Atractielomycetes(2) Soporidiobolales Nonidentified Nonidentified 2 2
Ustilagomycetes(2) Nonidentified Nonidentified Nonidentified 2 6 8
Nonidentified(5) Nonidentified Nonidentified Nonidentified 1 4 5
Marineclones (326)
350
300
250
200
150
100
50
0
Dotyideomycetes/Capnodiales/Capnodiaceae (1) Dotyideomycetes/Capnodiales/Davidiellaceae (1) Leotiomycetes/Helotiales/Dermateaceae, Bulgariaceae (2) Leotiomycetes/Helotiales/Non identified (2)
Sordariomycetes/Diaporthales/Non identified (4) Sordariomycetes/Hypocreales/Ophiocordycipitaceae (4) Eurotiomycetes/Mycocaliciales/Trichocomaceae (5) Sordariomycetes/Hypocreales/Incertae sedis (6) Zemp
oala
Carboner as
Mezquital
Cruz huan aca
xtle
Vice nte gu
errero Med
ia luna El r
abón
Tapachula Santa catar
ina
Bah ía de bander
as
Figure2.AscomycotadiversityinthesampledsitesidentifiedbyClass(sub-phyllum)/order/family.
Diversityestimation
MolecularOperationalTaxonomicalUnits(MOTUS) diver-sity was estimated using the Shannon Index, which gives a measure of both species numbers and the evenness of their abundanceaspresentedinTable4(Shannon,1948).Thevalue of the index ranges from low values (reduced species rich-nessandevenness)tohighvalues(extendedspeciesevenness and richness). In this work, the lowest diversity value was obtainedforMediaLunaandBahíadeBanderas,sampleswith a single MOTU identified in each (H=0). In contrast, the
45
40
35
30
25
20
15
10
5
0
Chytridiomycetes/Rhizophydiales/Rhizophydiaceae (4) Chytridiomycetes/Chytridiales/Chytridiaceae (4)
Chytridiomycetes/Spizellomycetales/Spizellomycetaceae (14) Non identified (33)
Chytridiomycetes/Chytridiales/Non identified (10) Chytridiomycetes/Spizellomycetales/Non identified (35) Zempo
ala
Carboner as
Mezquital
Cruz huanacaxtleVicen
te guerrero Medi a luna
El r abón
Tapa chula
Santa catar ina
Bahía de bander as
Figure3.ChytridiomycotadiversityinthesampledsitesidentifiedbyClass(sub-phyllum)/order/family.
highestdiversityvaluewasobtainedfortheZempoalalagoon (H=2.285), slightlyhigherthanthe totaldiversitybyfamily (H=2.130).
26 B.Valderramaetal./RevistaMexicanadeBiodiversidad87(2016)18–28
20
18
16
14
12
10
8
6
4
2
0
Non identified (15) Ustilaginomycetes/Non identified (8)
Mycrobotryomycetes/Leucosporidiales/Non identified (2) Tremellomycetes/Cystofilobasidales/Cystofilobasidiaceae (3) Mycrobotryomycetes/mitosporic Sporidiobolales (2)
Agaricomycetes/Polyporales/Non identified (13) Tremellomycetes/Filobasidiales/Filobasidieaceae (2) Zempoala Carbon
eras Mezqui
tal
Cruz huan acaxt
le
Vicente guerre ro
Media luna El r abón
Tapachula San
ta catar ina
Bahía de bander as
Figure4.BasidiomycotadiversityinthesampledsitesidentifiedbyClass(sub-phyllum)/order/family.
350
300
250
200
150
100
50
Rozzellida/Rozelliidae/Non identified (33) Eccrinales/Ichtyosporeae (1)
Matches to uncultivated clones from marine sources (326)
Choanoflagellidae/Acanthoecidae (12) Mucoromycotina/Mucorales (21)
Choanoflagellidae/Salpingoecidae (17) Zempoa
la
Carboner as
Mezquital
Cruz h uanacaxtle
Vicente gue rrero
Media luna
El r abón
Tapachu la
Santa catar ina
Bahía de band eras
Figure5.NonfungalmicrobialdiversityinthesampledsitesidentifiedbyClass(sub-phyllum)/order/family.
Discussion
Inaquaticenvironmentsorganicmatterdecompositionoccurs throughacomplexbutwelldefinedfungalsuccession(Barlocher &Kendrick,1974;Gessner, Thomas,Jean-Louis, &Chauvet, 1993).Themostcommonapproachusedforaquaticfungi diver-sity surveys involvesthe collection of organic material from natural sources such as plant debris or from artificial baits andthemicroscope-basedestimationofspeciesrichness.Using thisapproach, richness depends on the ability of the species inthecommunitytosporulate.Alternatively,molecular meth-ods may be applied for the identification of fungal species,
independentlyoftheirmetabolicstatusorlifecyclestage. Com-parative studies performed on decaying leaves indicate that bothapproachesarecomplementaryintheelucidationof pop-ulationcompositionanddynamics(Nikolcheva,Cockshutt,& Barlocher,2003).
Table4
Shannondiversityvaluescalculatedforthesampledsite.
Location Environment Npersite Spersite Hpersite
Zempoala Freshwatertemperatelagoon 171 25 2.285
Carboneras Brackishcoastallagoon 215 9 0.743
Mezquital Marinecoastallagoon 85 4 0.769
CruzdeHuanacaxtle PacificOceancoastline 64 8 1.749
VicenteGuerrero Dam 153 7 0.583
MediaLuna Brackishcoastallagoon 213 1 0
Elrabón Hypersalinecoastallagoon 241 3 0.148
Zacapulcp Mangroveswamp 361 8 0.906
SantaCatarina Artificialwaterreservoir 164 6 1.003
BahíadeBanderas PacificOceancoastline 326 1 0
Totalbyfamily 1,993 39 2.130
ofthesestudiesreferonlytocertaingroupsoffungi(yeasts,for example).
SmallsubunitribosomalDNAsequences(18SrDNA)have beenusedasmolecularmarkersforreconstructingfungal taxon-omy(Brunsetal.,1992;Hibbettetal.,2007;Jamesetal.,2006) andforthedescriptionoffungaldiversityinsoilsandwater bod-ies(Andersonetal.,2003;Huntetal.,2004;Monchyetal.,2011; Piquetetal.,2011).SmallsubunitrDNAsequenceshavebeen usedtoexplorebiologicaldiversityandspecializedsoftwarehas beendevelopedtodiscriminateamongprokaryoticand eukary-oticsequences(Bengtssonetal.,2011).18SrDNAsequencesare stillwidelyusedtoexploreenvironmentalsamples.Thesetof primersusedherewasdesignedfortheamplificationoffungal ribosomalDNA,inparticularaninternalfragmentof the18S ribosomalparticlegene betweenvariable regionsV4 andV8 (Borneman&Hartin,2000).Intheoriginalpapertheseprimers wereunabletoamplifyDNAisolatedfromorganismsotherthan fungiandoperationalspecificitywasdemonstratedbythe tar-getedamplificationoffungal18Sribosomalsequencesfromsoil samples(Andersonetal.,2003).
Species designation of non-cultured individuals based on molecularmarkers presentsthe intrinsicweakness of lacking a statistically sound method. In some cases the identifica-tion is based on the overall similarity of query sequences to referencesequencesinpublic databases in paired alignments usingarbitrarilydesignatedlimits.Inordertoenforcethe tax-onomical robustness of our work, we adapted the identity interval rank concept originally devised for the classifica-tionof plant-nodulatingbacterial species (Lloretet al.,2007; Martínez-Romero, Orme˜no-Orrillo, Rogel, López-López, & Martínez-Romero,2010).
Our approach is based on clustering of experimental sequences and a set of taxonomically classified reference sequencesencompassingknownfungalphylaandbasallineages. This gives enough information to reconstruct the taxonomic classificationofthe organisms.Sometimes clusteringpatterns were sensitive to the distribution of experimental sequences unlessmorereferencesequenceswereincluded.Inthesecases, we identified clusters of experimental entries lacking refer-ence sequences and used them to retrieve their best match inthe databases. Inclusionof thesenewreference sequences in the alignments settled tree topology. Each cluster was
taxonomicallyidentifiedusingtheclassificationofthereference sequenceslocated within, atdifferent levels,from phylumto family.
The sequencevariabilityof the 18S rDNAfragments sup-portstheclusteringreconstruction of ourreferencesequences assembledwithtaxonomicconsistency.Thisincludesthe iden-tificationof asubgroupof Chytridiomycetesbelongingtothe
Rozella genus,whichisknown tocluster separately(Hibbett etal.,2007).Itisimportanttonotethatwewereabletorecover sequencesfrom allfungal phyla, indicatinglittle bias for the collection procedureor the amplificationprimers used.From theentire collection,23.8%ofthe sequencescould be identi-fiedatthefamilylevel,48.5%attheorderleveland13%atthe class/subphylumlevel.
Theimportant roleof woodandleaflitterdegradation has beenascribedtoaquaticascomycetessincebasidiomycetesare scarce inwaterhabitats andotherorganismssuch asbacteria rarelyhavetheabilitytocompletelymineralizelignin(Simonis, Raja, &Shearer, 2008). Thus, it was not surprising that the Ascomycota was the most frequently recovered phylum and Sordariomycetesthemostfrequentlyrecoveredclass.The occur-renceofnon-identifiedascomyceteswasdocumentedinonly3 ofthesampledsitessupportingthatideathatthisphylumisthe best characterized eveninaquatic habitats. The second most abundantphylumdescribedinaquaticenvironmentsis Chytrid-iomycotaand,accordingly, itwasthe secondmostfrequently foundgroupforoursequencewith41%ofthesequences iden-tifiedatthefamilylevel.
28 B.Valderramaetal./RevistaMexicanadeBiodiversidad87(2016)18–28
Acknowledgments
ThisworkwaspartiallyfundedbygrantConacyt-Semarnat 2004-01-0100.WearegratefultoJoséLuisFernándezandDr. VíctorGonzálezfromCCG-UNAMforDNAsequencingand Prof.MichaelA.Pickardforcriticalreadingofthemanuscript.
References
Altschul,S.F.,Madden,T.L.,Schaffer,A.A.,Zhang,J.,Zhang,Z.,Miller,W., etal.(1997).GappedBLASTandPSI-BLAST:anewgenerationofprotein databasesearchprograms.NucleicAcidsResearch,25,3389–3402.
Anderson,I.C.,Campbell,C.D.,&Prosser,J.I.(2003).Potentialbiasoffungal 18SrDNAandinternaltranscribedspacerpolymerasechainreactionprimers forestimatingfungalbiodiversityinsoil.EnvironmentalMicrobiology,5, 36–47.
Barlocher,F.,&Kendrick,B.(1974).Dynamicsofthefungalpopulationon leavesinastream.JournalofEcology,62,761–791.
Bengtsson, J.,Eriksson, K. M.,Hartmann, M.,Wang, Z., Shenoy, B. D., Grelet,G.A.,etal.(2011).Metaxa:asoftwaretoolforautomated detec-tion and discrimination amongribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloro-plastsinmetagenomesandenvironmentalsequencingdatasets.AntonieVan Leeuwenhoek,100,471–475.
Borneman,J.,&Hartin,R.J.(2000).PCRprimersthatamplifyfungalrRNA genesfromenvironmentalsamples.AppliedandEnvironmental Microbiol-ogy,66,4356–4360.
Bruns,T.D.,Vilgalys,R.,Barns,S.M.,González,D.,Hibbett,D.S.,Lane,D. J.,etal.(1992).Evolutionaryrelationshipswithinfungi:analysesofnuclear smalsubunitrRNAsequences.MolecularPhylogeneticsandEvolution,1, 231–241.
Chapin,F.S.,Zavaleta,E.S.,Eviner,V.T.,Naylor,R.L.,Vitousek,P.M., Reynolds,H. L.,etal. (2000).Consequences ofchanging biodiversity. Nature,405,234–242.
Gessner,M.O.,Thomas,C.M.,Jean-Louis,A.M.,&Chauvet,E.(1993).
Stablesuccessionalpatternsofaquatichyphomycetesonleavesdecayingin asummercoolstream.MycologicalResearch,97,163–172.
González,M.,Hanlin,R.,Herrera,T.,&Ulloa,M.(2000).Fungicolonizing hair-baitsfromthreecoastalbeachesofMexico.Mycoscience,41,259–262.
González,M.,Hanlin,R.,&Ulloa,M.(2001).Achecklistofhighermarine fungiofMexico.Mycotaxon,80,241–253.
González,M.C.,&Chavarría,A.(2005).Somefreshwaterascomycetesfrom Mexico.Mycotaxon,91,315–322.
Guzmán,G.(1998).InventoryingthefungiofMexico.Biodiversityand Con-servation,7,369–384.
Hawksworth,D.L.(1991).Thefungaldimensionofbiodiversity:magnitude, significance,andconservation.MycologicalResearch,95,641–655.
Hawksworth,D.L.(2001).Themagnitudeoffungaldiversity:the1.5million speciesestimaterevisited.MycologicalResearch,105,1422–1432.
Heredia,G.,Reyes,M.,Arias,R.M.,Mena-Portales,J.,&Mercado-Sierra,A. (2004).Adicionesalconocimientodeladiversidaddeloshongos conidi-alesdelbosquemesófilodemonta˜nadelestadodeVeracruz.ActaBotanica Mexicana,66,1–22.
Hibbett,D.S.,Binder,M.,Bischoff,J.F.,Blackwell,M.,Cannon,P.F.,Eriksson, O.E.,etal.(2007).Ahigher-levelphylogeneticclassificationoftheFungi. MycologicalResearch,111,509–547.
Hunt,J.,Boddy,L.,Randerson,P.F.,&Rogers,H.J.(2004).Anevaluationof 18SrDNAapproachesforthestudyoffungaldiversityingrasslandsoils. MicrobialEcology,47,385–395.
James,T.Y.,Kauff,F.,Schoch,C.L.,Matheny,P.B.,Hofstetter,V.,Cox,C.J., etal.(2006).Reconstructingtheearlyevolutionoffungiusingasix-gene phylogeny.Nature,443,818–822.
Kimura,N.(2006).Metagenomics:accesstounculturablemicrobesinthe envi-ronment.MicrobesandEnvironments,21,201–215.
Klaubauf,S.,Inselsbacher,E.,Zechmeister-Boltenstern,S.,Wanek,W., Gotts-berger, R., Strauss, J., et al. (2010). Molecular diversity of fungal communitiesinagriculturalsoilsfromLowerAustria.FungalDiversity, 44,65–75.
Larkin,M.A.,Blackshields,G.,Brown,N.P.,Chenna,R.,McGettigan,P. A.,McWilliam,H.,etal.(2007).ClustalWandClustalXversion2.0. Bioinformatics(Oxford,England),23,2947–2948.
LeCalvez,T.,Burgaud,G.,Mahé,S.,Barbier,G.,&Vandenkoornhuyse,P. (2009).Fungaldiversityindeep-seahydrothermalecosystems.Appliedand EnvironmentalMicrobiology,75,6415–6421.
Lloret,L.,Orme˜no-Orrillo,E.,Rincón-Rosales,R.,Martínez-Romero,J., Rogel-Hernández,M.A.,&Martínez-Romero,E.(2007).Ensifermexicanumsp. nov.anewspeciesnodulatingAcaciaangustissima(Mill.)KuitzeinMexico. SystematicandAppliedMicrobiology,30,280–290.
Martínez-Romero,J.C.,Orme˜no-Orrillo,E.,Rogel,M.A.,López-López,A., & Martínez-Romero,E.(2010).Trendsinrhizobialevolutionandsome taxonomicremarks.InP.Pontarotti(Ed.),Evolutionarybiology–concepts, molecularandmorphologicalevolution(pp.301–315).Berlin:Springer.
Monchy,S.,Sanciu,G.,Jobard,M.,Rasconi,S.,Gerphagnon,M.,Chabe,M., etal.(2011).Exploringandquantifyingfungaldiversityinfreshwaterlake ecosystemsusingrDNAcloning/sequencingandSSUtagpyrosequencing. EnvironmentalMicrobiology,13,1433–1453.
Moon-vanderStaay,S.Y.,DeWachter,R.,&Vaulot,D.(2001).Oceanic18S rDNAsequencesfrompicoplanktonrevealunsuspectedeukaryoticdiversity. Nature,409,607–610.
Mueller,G.,&Schmit,J.(2007).Fungalbiodiversity:whatdoweknow?What canwepredict?BiodiversityandConservation,16,1–5.
Mueller,G.M.,Bills,G.F.,&Foster,M.S.(2004).Biodiversityoffungi inven-toryandmonitoringmethods(Vol.1)Burlington,MA:ElsevierAcademic Press.
Nikolcheva, L.G.,Cockshutt, A.M.,& Barlocher,F.(2003).Determining diversityoffreshwaterfungiondecayingleaves:comparisonoftraditional andmolecularapproaches.AppliedandEnvironmentalMicrobiology,69, 2548–2554.
Piquet,A.M.,Bolhuis,H.,Meredith,M.P.,&Buma,A.G.(2011).Shiftsin coastalAntarcticmarinemicrobialcommunitiesduringandaftermelt water-relatedsurfacestratification.FEMSMicrobiologyEcology,76,413–427.
Saitou,N.,&Nei,M.(1987).Theneighbor-joiningmethod:anewmethod forreconstructingphylogenetictrees.MolecularBiologyandEvolution,4, 406–425.
Shannon,C.E.(1948).Amathematicaltheoryofcommunication.BellSystem TechnicalJournal,27,379–423.
Shearer,C.,Descals,E.,Kohlmeyer,B.,Kohlmeyer,J.,Marvanova,L.,Padgett, D.,etal.(2007).Fungalbiodiversityinaquatichabitats.Biodiversityand Conservation,16,49–67.
Simonis,J.L.,Raja,H.A.,&Shearer,C.A.(2008).Extracellularenzymesand softrotdecay:areascomycetesimportantdegradersinfreshwater?Fungal Diversity,31,135–146.
Tamura,K.,Dudley,J.,Nei,M.,&Kumar,S.(2007).MEGA4:Molecular Evolu-tionaryGeneticsAnalysis(MEGA)softwareversion4.0.MolecularBiology andEvolution,24,1596–1599.
Torsvik,V.,&Ovreas,L.(2002).Microbialdiversityandfunctioninsoil:from genestoecosystems.CurrentOpinioninMicrobiology,5,240–245.
Vainio,E.J.,&Hantula,J.(2000).Directanalysisofwood-inhabitingfungi usingdenaturinggradientgelelectrophoresisofamplifiedribosomalDNA. MycologicalResearch,104,927–936.
VanderAuwera,G.,DeBaere,R.,VandePeer,Y.,DeRijk,P.,VandenBroeck, I.,&DeWachter,R.(1995).ThephylogenyoftheHyphochytriomycota asdeducedfromribosomalRNAsequencesofHyphochytriumcatenoides. MolecularBiologyandEvolution,12,671–678.