Pan genome of the phytoplankton Emiliania underpins its global distribution
Texto completo
Figure
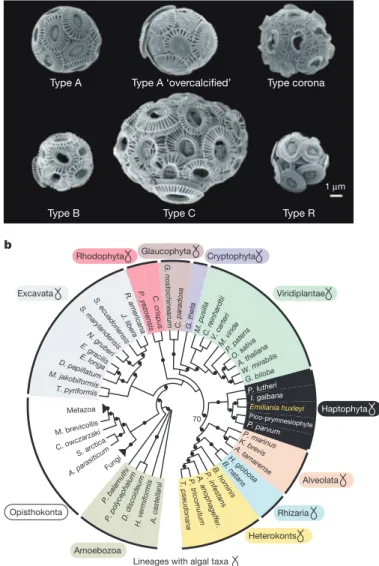
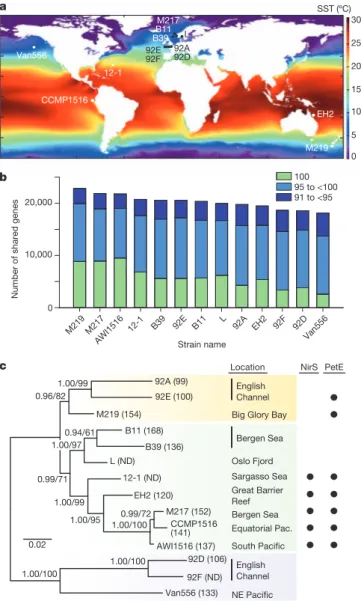
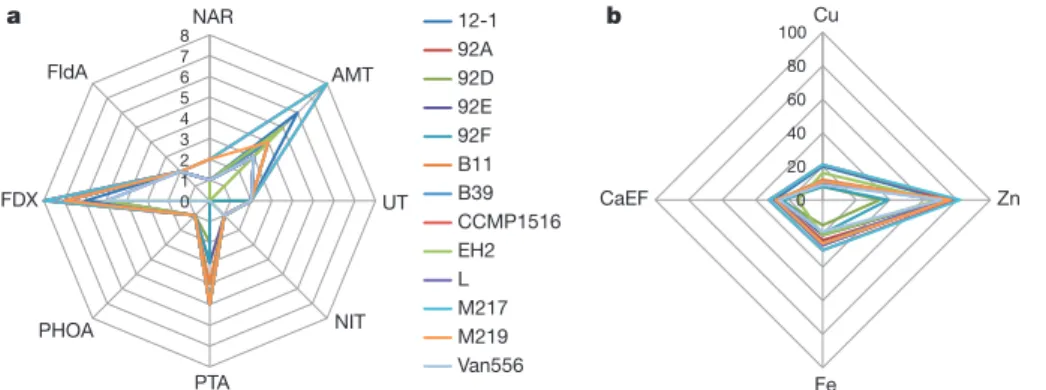
Documento similar
In the “big picture” perspective of the recent years that we have described in Brazil, Spain, Portugal and Puerto Rico there are some similarities and important differences,
In the second part of this thesis, the molecular and structural characterization of Ad5/FC31 light particles revealed that these particles lack genome and
Besides transcription factors, several chromatin features have been shown to regulate the transcriptional activity of a promoter, like the nucleosomal configuration
No obstante, como esta enfermedad afecta a cada persona de manera diferente, no todas las opciones de cuidado y tratamiento pueden ser apropiadas para cada individuo.. La forma
This stage performs five consecutive tasks: i) CALs are extended with additional information from the reference genome; ii) extended CALs which are close to each other in the
Díaz Soto has raised the point about banning religious garb in the ―public space.‖ He states, ―for example, in most Spanish public Universities, there is a Catholic chapel
(A) Distribution of the total pangenome after core genome multilocus sequence typing (cgMLST) analysis in core (genes present it at least 95% of the isolates; 7.6%), accessory
Results: In order to determine the ability of the host genome to control the diversity and composition of microbial communities in healthy pigs, we undertook genome-wide