TítuloModels of landscape evolution and the survival of Palaeoforms
Texto completo
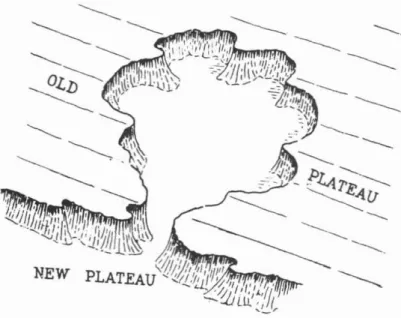
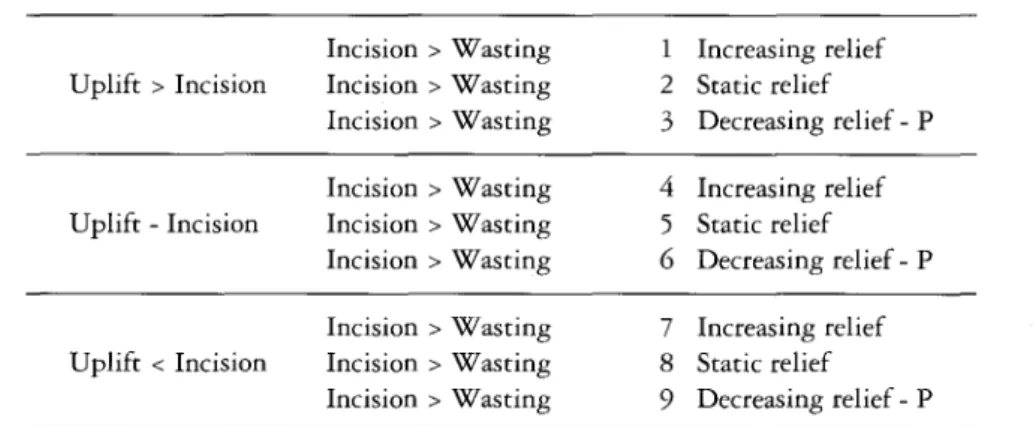
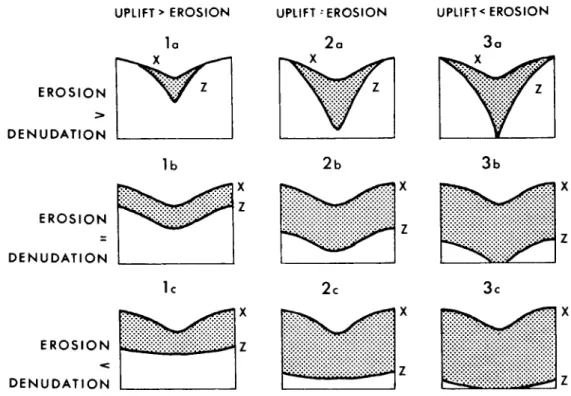
Figure


Documento similar
When the evolution of the drop takes place at constant angular momentum, as is the case when the drop is mechanically isolated, and axial symmetry is imposed, we always converge to
EP-GRN arose by co-option (Nanog, Sox2, Fgf4), duplication (Pou2-r) and the appearance of novel genes (Dppas, Utf1), as well as new regulatory interactions that recruited
However, given the present scenario of scarce allosteric-site data, we decided to perform a large-scale analysis of protein ligand-binding pockets, as these
Globular clusters are ideal laboratories for testing theories of stellar evolution, the chemical evolution of the Universe and the dynamics of N-body systems' They are
Overall Rate of Evolution and Information Scores Phylogenetic information scores are calculated assuming an overall rate of evolution (that of the full data set comprising
In Section 2 we consider abstract evolution equations with memory terms and prove that the null and memory-type null controllability properties are equivalent to certain
Ancestral sequence reconstruction (ASR) and resurrection (i.e. functional expression in a heterologous host) allows enzymes with different properties to be disclosed while its
(C) lncRNAs can represent a bridge between the many layers of epigenetic gene regulation, interacting with, for example, histone modifiers or serving as ceRNAs for miRNAs; (D)