Role of cerebral dopamine neurotrophic factor in endoplasmic reticulum stress
Texto completo
Figure
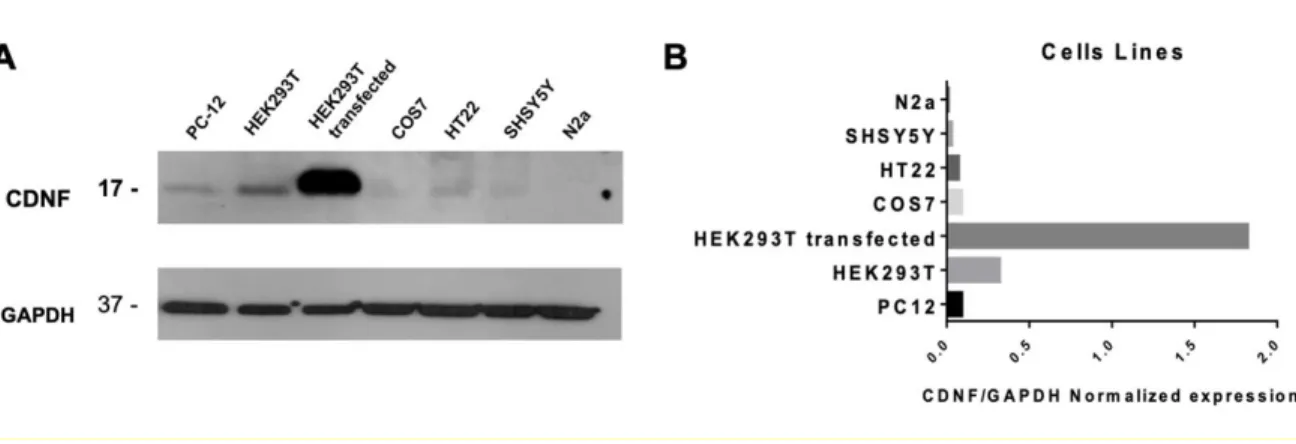
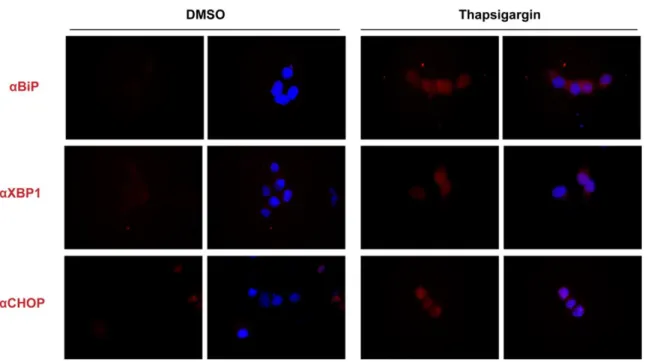
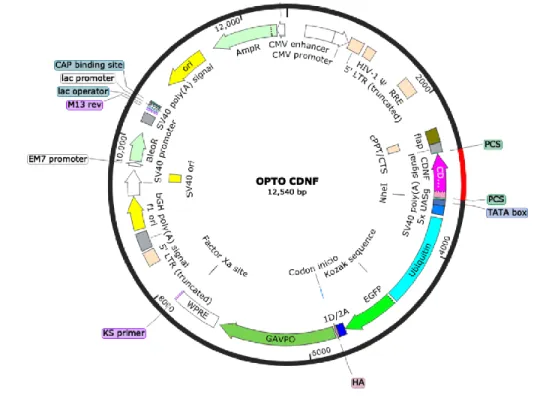
Documento similar
(A) HIF1 binding locations common to at least two out of four different ChIP-chip studies in HeLa, HepG2, MCF-7 and U87 cells (left), mammalian sequence conservation of the HIF
Glial Cell Line-Derived Neurotrophic Factor Promotes the Arborization of Cultured Striatal Neurons Through the p42/p44 Mitogen Activated Protein Kinase Pathway..
Inoue, Y., et al., Role of hepatocyte nuclear factor 4alpha in control of blood coagulation factor gene expression.. Safdar, H., et al., Modulation of mouse
At the same time, ZEB1 can switch from a transcriptional repressor to an activator by binding to p300/CBP suggesting another potential mechanism by which ZEB1 regulates
In addition, accumulating evidence in human lesion patients on the Val66Met BDNF polymorphism and cognitive function indicates that the Met allele exerts a protective effect
Our preliminary data on patient-derived MES GBM cells, in which we observed a downregulation of the MES signature upon FOSL1 knockdown (Figure R.20A-B), is quite convincing that
En este sentido, es interesante mencionar que la expresión de TCFL5 aparece en las fases finales de diferenciación a línea germinal coincidente con una disminución de los genes
In situ hybridization in late third instar wing discs of genes which expression levels change comparing 756-Gal4/UAS-sal vs 756-Gal4/UAS-GFP (UAS) or wild type vs Df(2L) 32FP5