A Systems Approach Uncovers Restrictions for Signal Interactions Regulating Genome wide Responses to Nutritional Cues in Arabidopsis
Texto completo
Figure
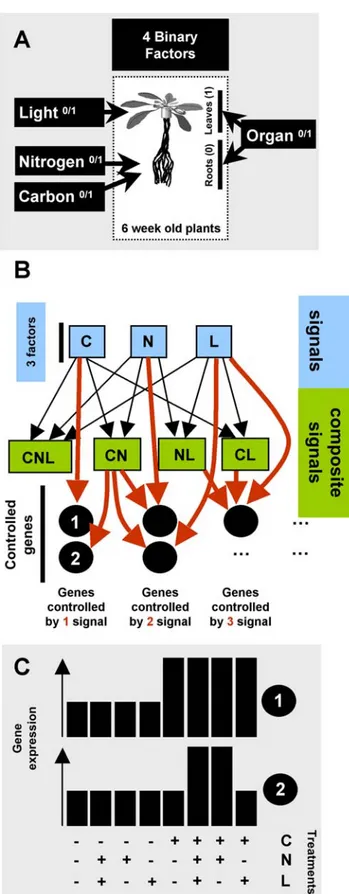
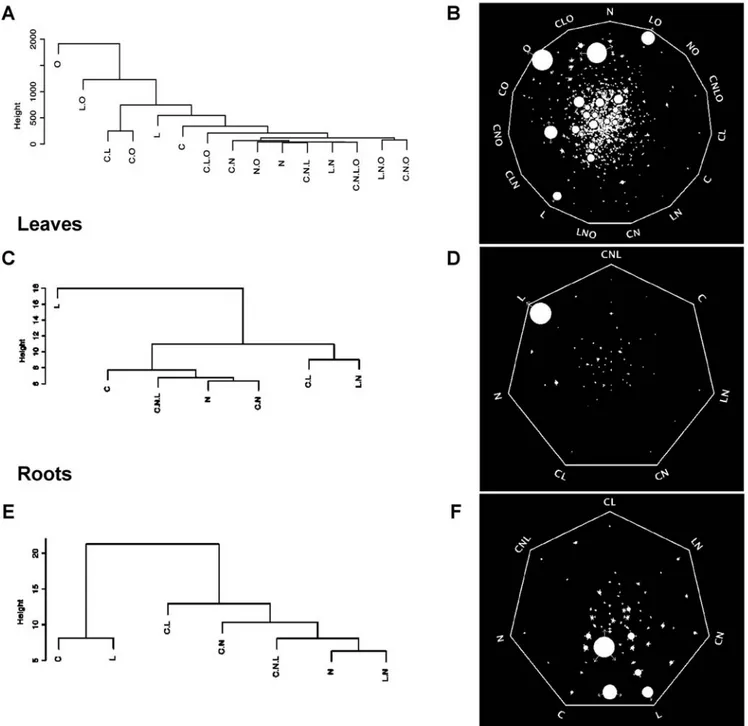
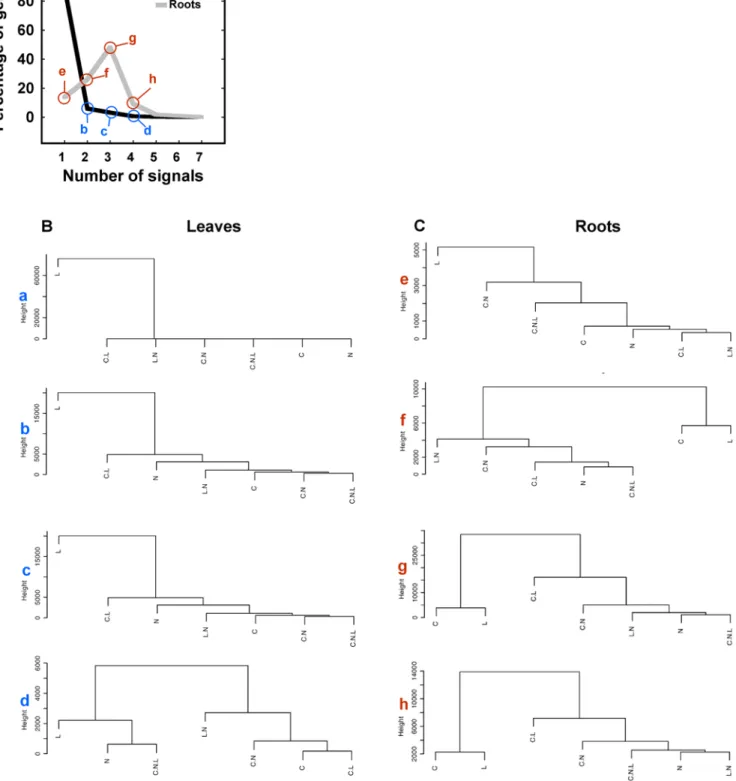
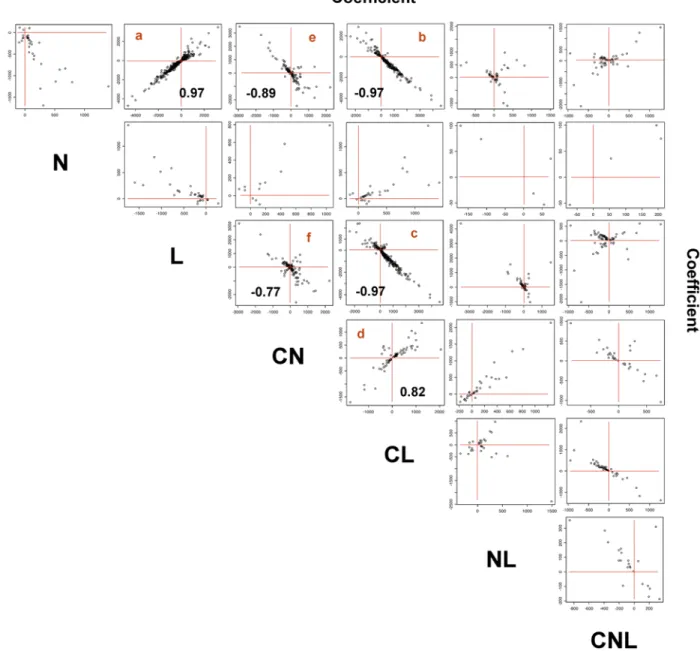
Documento similar
In the present study, gene expression of candidate genes for fatty acid composition in muscle showed sex-dimorphism and breed effects, and gene co-expression in different
The greater C S values obtained for G/N-CFs prove that the incorporation of graphene in the nitrogen doped carbon electrode improves its conduction properties and, as a
The analysis of differential gene expression resulted in 39 differentially expressed genes (DEGs) between mites exposed to tomato and those exposed to pepper leaves in four
Results: In order to determine the ability of the host genome to control the diversity and composition of microbial communities in healthy pigs, we undertook genome-wide
In order to identify differential gene expression profiles that could be used as markers of response to anti-TNF treatment, we compared the relative expression of TLR2, TNF,
In conclusion, our STAT-inhibitory studies of C01L_F03, STATTIC and STX-0119 and our previous revelation of STAT cross-binding of a pre-selection of known STAT3 inhibitors
In this study we performed a detailed genome-wide lin- kage analysis using MOD scores and 6 families affected by different CHD to test for a common genetic background among
Discarding the role of LrpC in global gene expression, the in vivo localization of GFP-LrpC during vegetative role was addressed, in order to obtain a hint of the possible